Urine is Not Sterile: the Bladder Microbiome
Traditionally the healthy bladder has been believed to be sterile. This was based on a lack of bacterial growth when attempting to culture urine on media. The discovery of the microbiome, the bacteria and other microbes that live in and on our bodies, has upended this dogma. Culture-independent, molecular methods of characterizing the microbiome have revealed that even in healthy individuals with normal bladders and negative urine cultures, the urine contains numerous bacteria. We helped demonstrate this in Groah et al.. This paper made a number of interesting observations. For instance, men and women had different urine microbiomes:
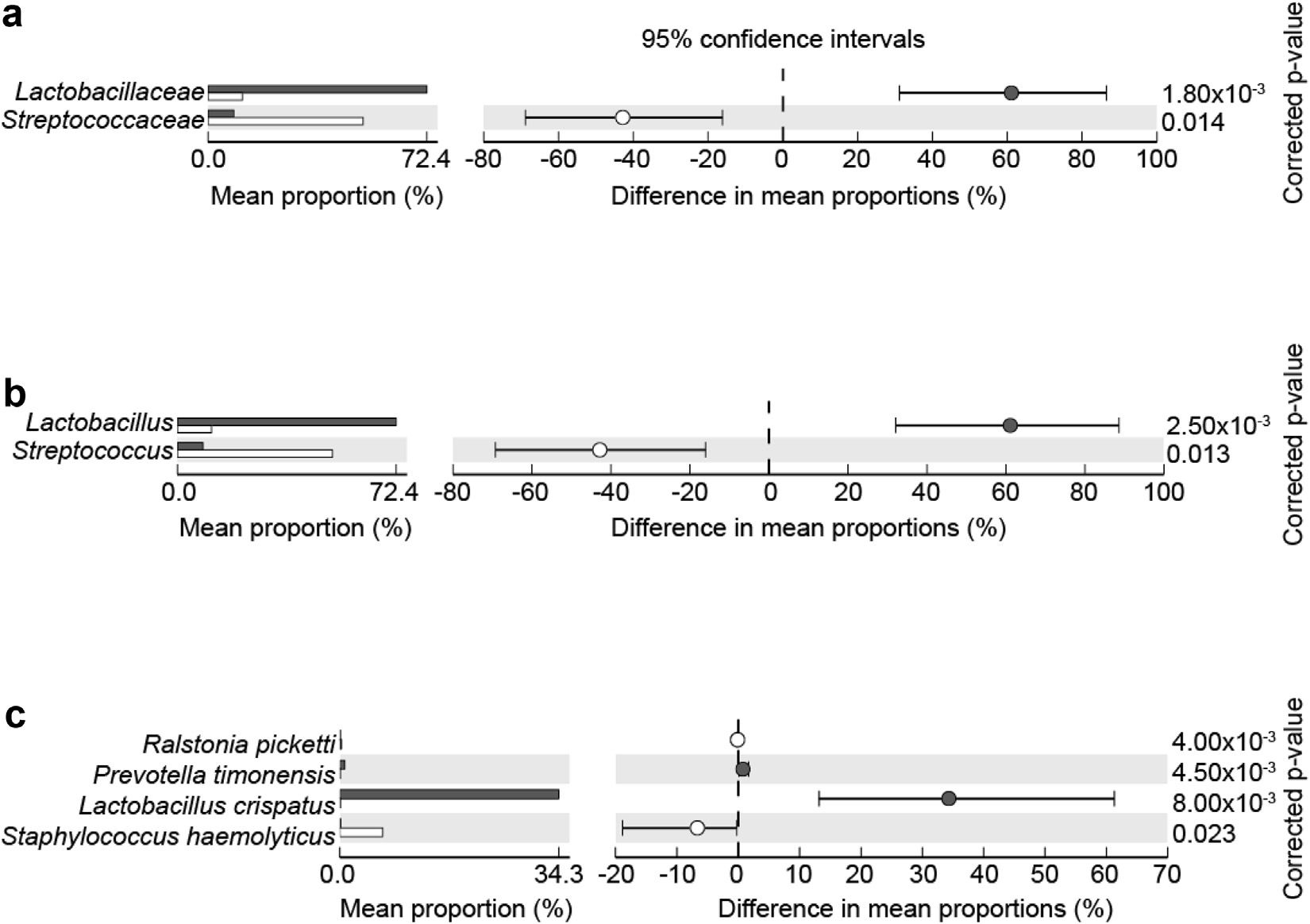
Significant differences in urine microbiomes of all females (gray bars) vs. all males (open bars) at family (a), genus (b), and species (c) levels.
We also noted that the urine microbiomes differed between patients with normal vs. neuropathic bladders:
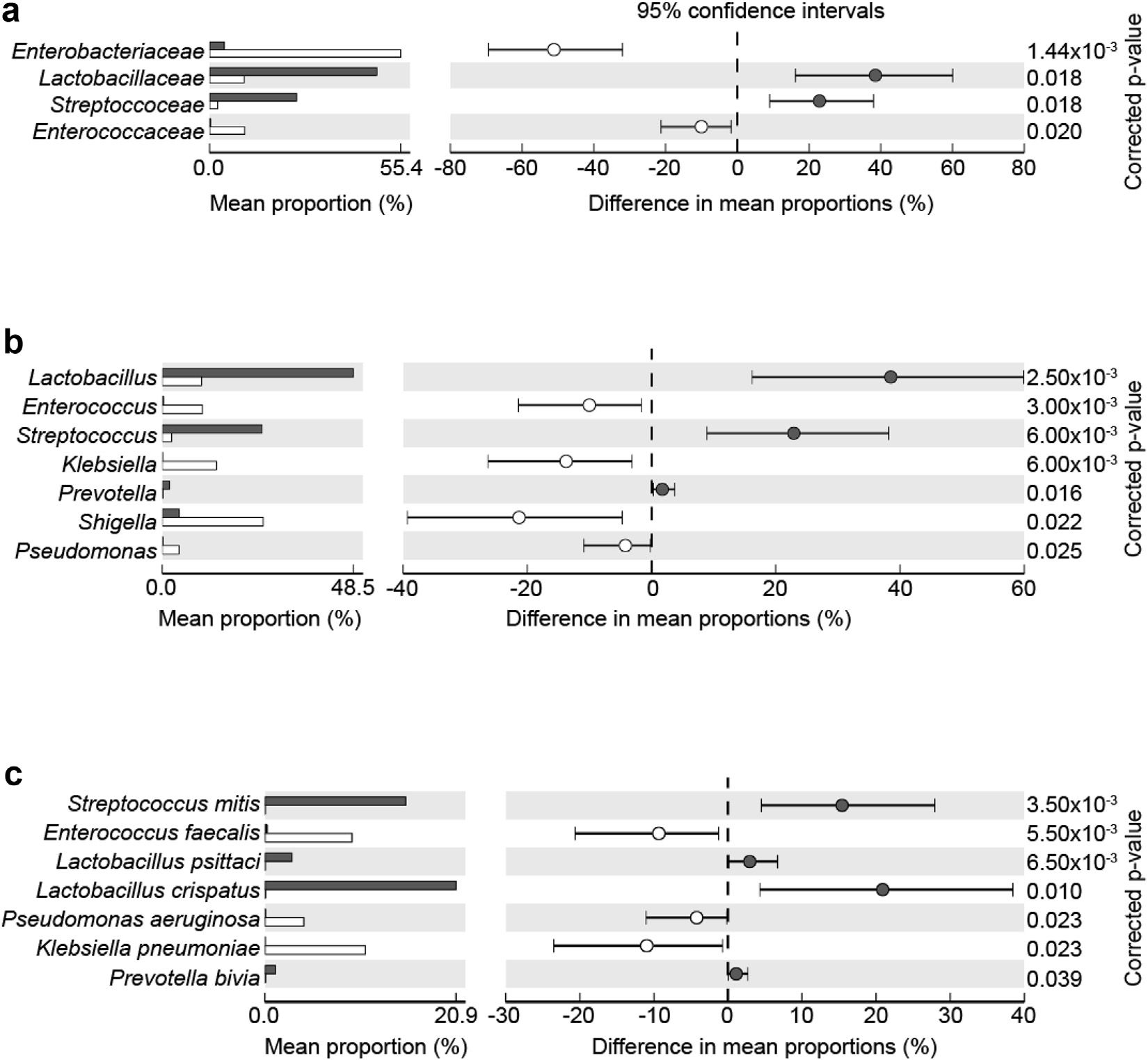
Significant differences in group urine microbiome analyses of non-neuropathic bladder (gray bars) vs neuropathic bladder (open bars) subjects at family (a), genus (b) and species (c) levels.
We also observed that neuropathic bladder patients who either had a suprapubic tube or relied upon intermittent urethral catheterization to empty their bladders tended to have more Enterobacteriaceae and less Lactobacillus in their urine than patients with normal bladders and patients with neuropathic bladder who were not catheterization-dependent:
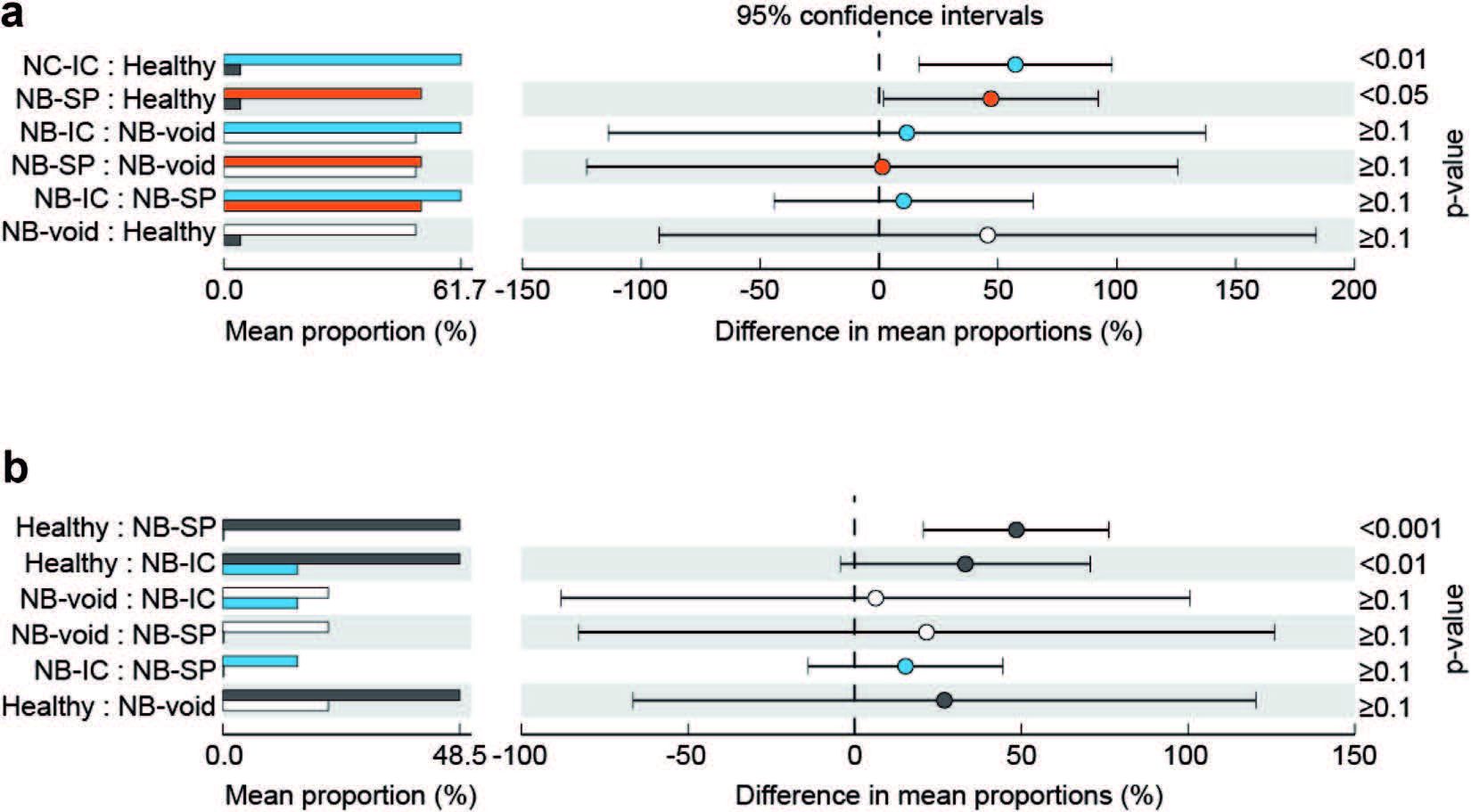
Significant differences in Enterobacteriaceae (a) and Lactobacillaceae (b) by bladder function and management method in neuropathic bladder subjects using intermittent catheterization (blue bars), neuropathic bladder subjects using suprapubic tubes (red bars), non-neuropathic bladder subjects (black bars) and neuropathic bladder patients who voided (open bars).
We were surprised to find that there was no apparent relationship between microbial diversity and pyuria:
Regardless, our findings contribute to the burgeoning literature confirming that the urine is not sterile. These observations have important implications for bladder health and disease; it may be possible to manipulate the microbiome to affect outcomes of bladder-related conditions.
Approximately half of all girls and women and some boys and men will experience at least one urinary tract infection (UTI) during their lifetime. UTI are the most common hospital-associated infection, and can lead to death in critically ill patients. Effective therapy is based on antibiotics, but bacterial resistance is an ongoing issue for management of UTI.
An unexpected source of antibiotic resistance among bacterial uropathogens may be the general stress response. This pathway is activated by a number of environmental stressors, including antibiotics, starvation, and disinfectants. In collaboration with A.C. Matin‘s laboratory, we demonstrated the in vivo mechanisms by which the general stress response imbues bacteria with resistance to specific antibiotics. Specifically, the general response gene rpoS seems to confer resistance of uropathogenic E. coli to gentamicin:

Panel A shows that rpoS-deficient uropathogenic E. coli (delta rpoS) are more susceptible to gentamicin treatment of infected mice compared to wild type uropathogenic E. coli (“wt”). Panel B shows that giving uropathogenic E. coli-infected mice the reactive oxygen species scavenger N-acetylcysteine (“NAC”) rescues rpoS-deficient uropathogenic E. coli (delta rpoS) from the bactericidal effects of gentamicin (“Gm”). From Wang et al.
We have worked closely with colleagues at Stanford University to develop new aminoglycoside-inspired antibiotics that not only feature less ototoxicity (inner ear damage is a well-known complication of use of these antibiotics), but also exhibit preserved antibacterial activity in the mouse model of UTI:
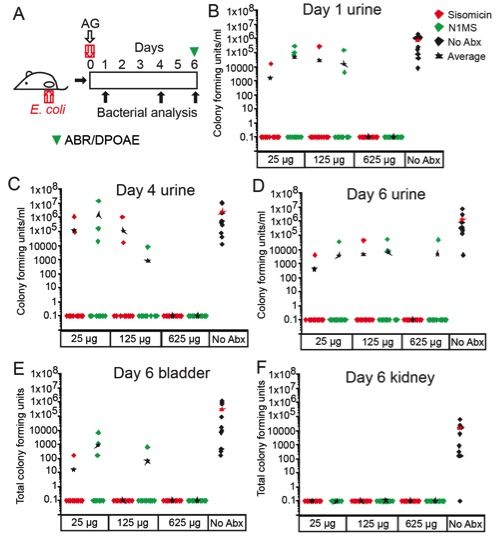
Panel A shows a schematic of the experimental design. Mice are transurethrally inoculated with uropathogenic E. coli and given aminoglycoside (“AG”) antibiotics on the same day. On days 1, 4, and 6 post-infection, urine, bladder, and kidney levels of bacterial infection were measured, and on day 6 post-infection hearing function was also measured (ABR/DPOAE). Panels B-E demonstrate that higher levels of both sisomicin (the parent aminoglycoside compound) and the non-ototoxic N1MS derivative antibiotic both clear bacteria better than no antibiotics. From Huth et al.
Below is video footage from our Journal of Visualized Experiments article on the mouse transurethral catetherization-based UTI model:
Urinary Tract Co-Infections
We are also interested in urinary tract co-infections, an understudied area of UTI biology. Urinary tract co-infections can be polymicrobial, in which more than one bacterial uropathogen have taken hold in host urinary tract tissues. Other urinary tract co-infections can be cross-kingdom, wherein eukaryotic uropathogens (i.e., Candida species or Schistosoma haematobium) have infected the same host urinary tracts as their bacterial brethren. In all of these settings, it is plausible that the interactions among multiple uropathogens and their hosts result in unique pathobiology.
For instance, numerous studies have observed that people with urogenital schistosomiasis (infection by Schistosoma haematobium worms) have higher rates of bacterial UTI than expected. It is unclear whether this purported association is a true biological link, or merely detection bias. The association between urogenital schistosomiasis and bacterial UTI susceptibility could be subject to detection bias; since people with S. haematobium infection often have urinary symptoms such as hematuria, they may come to urologic attention and undergo bacterial urine cultures more frequently than people without urogenital schistosomiasis.
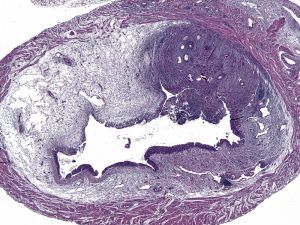
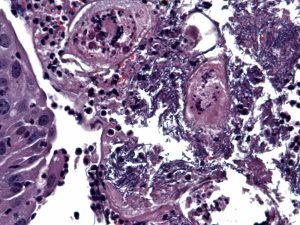
Even if there is a true biological connection between urogenital schistosomiasis and susceptibility to bacterial UTI, the associated mechanisms are not understood. Hypothesized mechanisms include urinary stasis from fibrotic obstruction and host immunomodulation by the parasite. We published the first mouse model of S. haematobium-bacterial uropathogen co-infection (Hsieh et al., Infection and Immunity, 2014). In this model, a single bladder exposure to S. haematobium eggs triggers granuloma formation that can be seen by ultrasound and histology of bladder sections. This resembles human bladder schistosomiasis:

Co-exposure of the mouse bladder to S. haematobium eggs and bacterial uropathogens also triggered production, a cytokine classically associated with schistosomiasis:
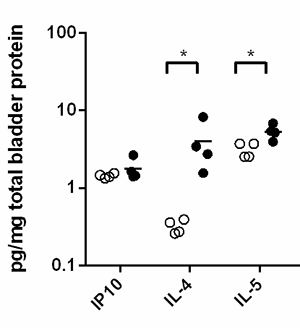
Bladder cytokines were measured by Luminex assays after 2 days of UTI89 infection. Co-exposed (S. haematobium eggs and uropathogenic E. coli strain UTI89) mice are denoted by closed circles, mono-infected mice (UTI89 only) by open circles. (n=4/group). Adapted from Hsieh et al., Infection and Immunity, 2014
Importantly, we found that a single exposure to parasite eggs also makes BALB/c mice susceptible to bacterial UTI when they are otherwise resistant:
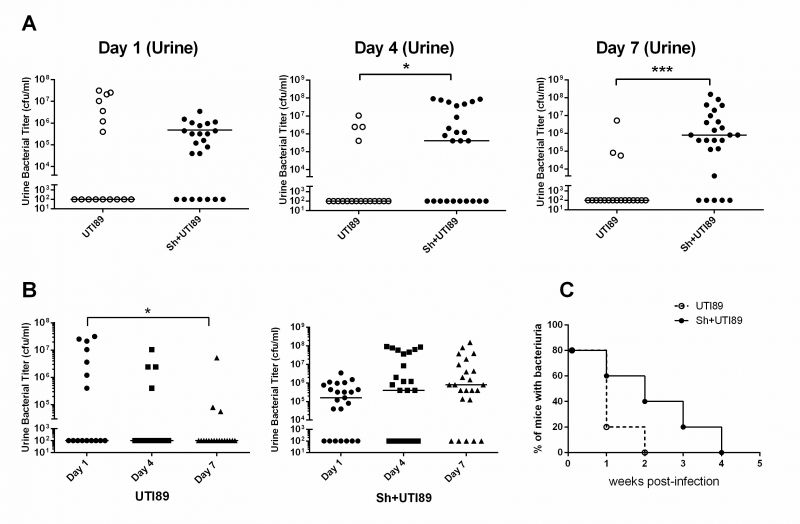
3000 S. haematobium eggs (Sh+UTI89; closed circles) or control vehicle (UTI89; open circles) were injected into the bladder walls of mice (A-C). 7 days later, all mice were transurethrally inoculated with 10E8 cfu UTI89. (A-B) Urine was collected for bacterial counts 1, 4, and 7 days post-UTI89 infection. Data shown were compiled from four independent experiments. (C) Urine bacterial titers in co-exposed mice were measured weekly (n=5). Bacterial cfu differences between groups were compared using Mann-Whitney U tests. *p<0.05, **, p<0.01, ***, p<0.001; horizontal bars (A-C) indicate median values. Adapted from Hsieh et al., Infection and Immunity, 2014
On occasion filamentous-appearing E. coli cells can be seen in the vicinity of eggs (ovoid objects below) within bladder granulomata.
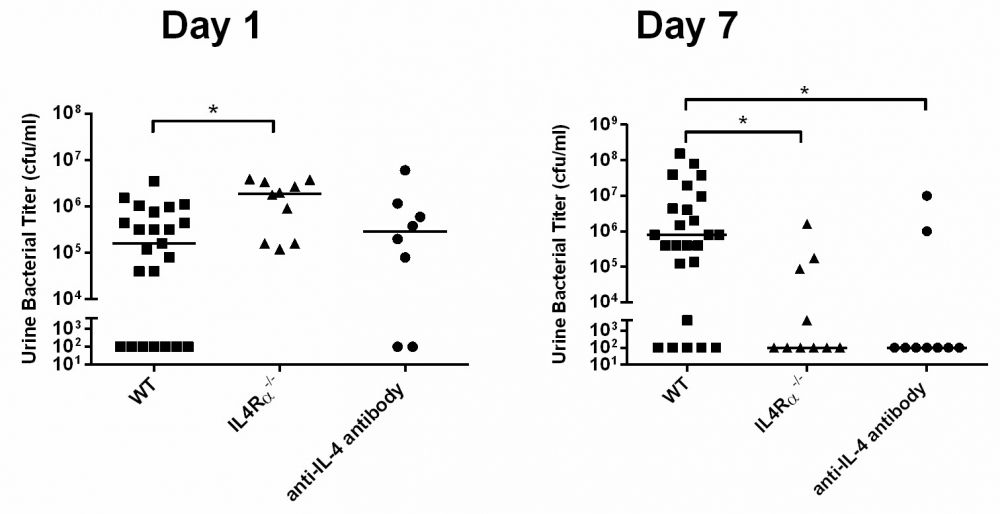
Given the prominence of bladder IL-4 expression after exposure to S. haematobium eggs, we next asked whether this cytokine plays a role in susceptibility to bacterial UTI. Indeed, we found that ablation of IL-4 receptor alpha (IL-4Rα) signaling and neutralization of IL-4 restored the baseline resistance of BALB/c to bacterial UTI despite prior exposure to S. haematobium eggs:
In the course of characterizing the leukocytic infiltrate of bladders in our model, we found that numbers of NKT cells were decreased in co-exposed versus bacterial mono-infected bladders:
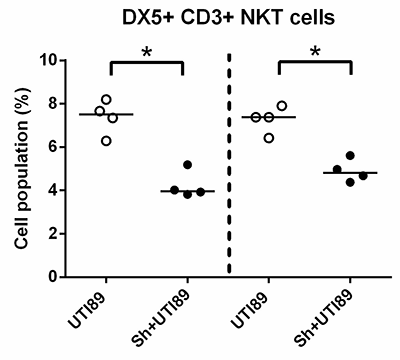
Flow cytometric analyses of single cell suspensions isolated from bacterial mono-infected and co-exposed bladders demonstrated the presence of NKT cells (DX5+CD3+). Co-exposed mice are denoted by “Sh+UTI89” (closed circles), mono-infected mice by “UTI89” (open circles). Adapted from Hsieh et al., Infection and Immunity, 2014
Given that schistosome-induced, non-NKT cell leukocyte infiltration may dilute NKT cell numbers in the bladders of co-exposed mice without exerting a specific functional effect on these cells, we next examined NKT cell biology on a per-cell basis. Invariant NKT (iNKT) cells from co-exposed mice expressed less interferon-γ (IFN-γ) per cell compared to those from mice infected with UTI alone:
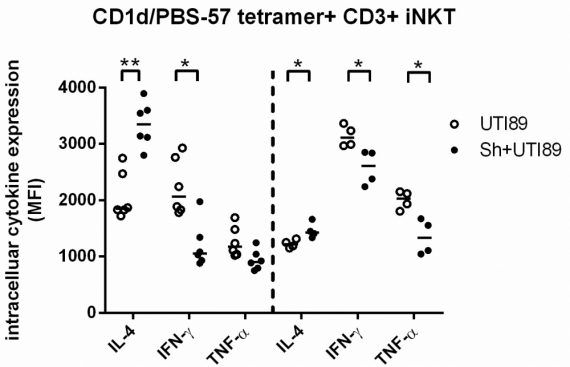
Intracellular cytokine staining of bladder iNKT cells from mono- and co-exposed mice (n=6/group). Data from two independent experiments are shown and separated by vertical dotted line. Adapted from Hsieh et al., Infection and Immunity, 2014
Since NKT cell activation appeared to be compromised, we then examined the biology of CD1d, a crucial antigen-presenting molecule for NKT cells, and any associated links with IL-4. Accordingly, we found that co-exposure resulted in lower CD1d expression in bladder antigen-presenting cells (APC) than in bacterial UTI alone in an IL-4Rα-dependent fashion:

CD1d expression on CD11c+CD3-B220- DC cells and F4/80+CD11b+SiglecF- macrophages was analyzed in mono-infected (“WT UTI89”, open circles), co-exposed (“WT Sh+UTI89”, closed circles) wild type and co-exposed IL4Rα−/− (“IL4Rα−/− Sh+UTI89”, open triangles) mice. Data are representative of one of two independent experiments. “MFI” denotes median fluorescence intensity. Treatment groups were compared using ANOVA followed by post hoc pairwise comparisons. *p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0005; horizontal bars indicate median values. Adapted from Hsieh et al., Infection and Immunity, 2014
Finally, co-exposed mice were protected from prolonged bacterial infection by administration of alpha-galactosylceramide, an iNKT cell agonist:
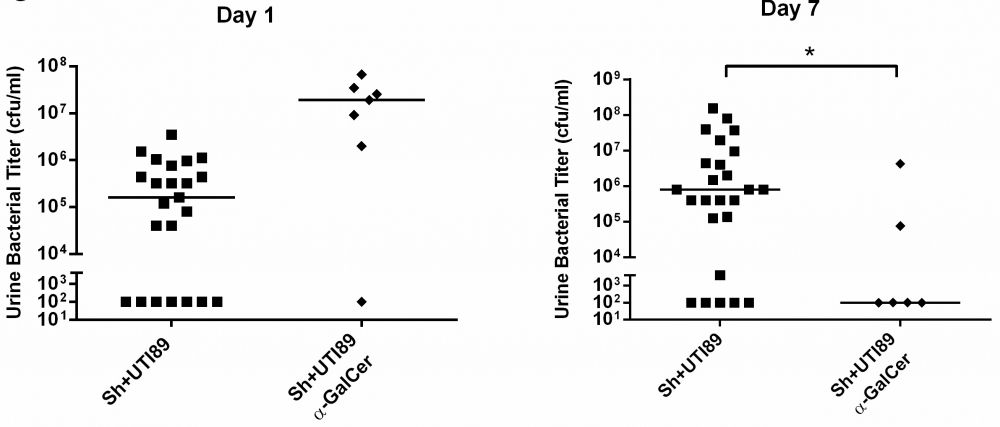
Effect of α-GalCer on UTI89 clearance in co-exposed mice. Co-exposed mice were given intraperitoneal α-GalCer (2 μg, “Sh+UTI89+α-GalCer”) on Day 0, 2, and 5 (or no α-GalCer) after UTI89 infection. Urine bacterial titers were examined at 1 and 7 days post-UTI89 infection. Data were compiled from two to four independent experiments. *p<0.05, **, p<0.01, ***, p<0.001; horizontal bars indicate median values. “MFI” denotes median fluorescence intensity. Co-exposed mice are denoted by “Sh+UTI89” (closed circles). Student’s t tests were used to compare values between treatment groups. Adapted from Hsieh et al., Infection and Immunity, 2014
Given that S. mansoni eggs produce copious amounts of the interleukin-4 inducing principle of S. mansoni eggs (IPSE), our theoretical model is that S. haematobium eggs, like their S. mansoni counterparts, induce basophil and/or T cell production of IL-4. We believe IL-4 acts upon bladder APC (macrophages and DC) to decrease their expression of CD1d and MHC Class II. In turn, this results in suboptimal bladder NKT cell activation for promoting clearance of bacterial urinary tract infection, a decreased IFN-γ:IL-4 ratio among bladder iNKT cells in particular, and prolonged bacteriuria:
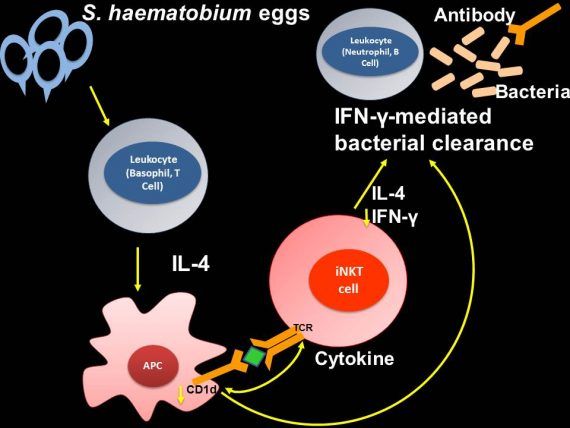
These findings underscore the potential benefits of first clearing helminth infections prior to treating bacterial co-infections in affected individuals. In addition, our data indicate that, in some people with helminth infection and antibiotic-refractory or –resistant bacterial co-infections, NKT cell-directed therapies may be non-antibiotic-based alternatives.
Developing Better UTI Diagnostics
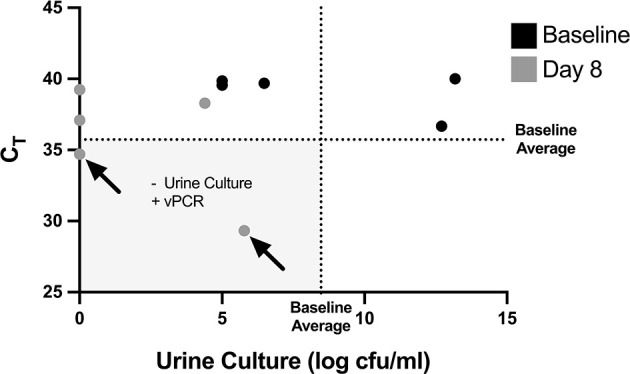
The current gold standard for diagnosis of urinary tract infections (UTI) currently depends upon growing bacteria in culture. Final identification of bacterial species and their antibiotic susceptibility can take days, which delays optimal care of patient with UTI. In addition, urine cultures have a significant false negative rate due to many reasons, including the presence of fastidious organisms that do not grow well in culture, and masking of bacterial growth following recent antibiotic exposure. This has led to the emergence of molecular methods to diagnose UTI, which rely upon PCR or sequencing to identify bacterial DNA. These methods have in turn been criticized because they may yield false positive results by identifying DNA from dead bacteria. We have developed propidium monoazide-based PCR methods for amplifying DNA from live bacteria in urine. Our results suggest that non-PMA bound DNA from live bacteria can be present in urine, even after antibiotic treatment. This indicates that viable but non-culturable E. coli can be present following treatment of UTI, and may explain why some patients have persistent symptoms but negative urine cultures following UTI treatment.